GraphDINO with Deep Graph Neural Networks
The brain is structured into various areas that each contain different types of neurons. Clustering of such cortical neurons by their shape (including its branching structure) has a long tradition in biology research. It is known that the shape of a neuron is tightly connected to its function in the brain. In the last couple of years, the amount of data has risen massively such that manual inspection is no longer feasible. We are thus interested in purely data-driven methods to cluster such neurons. Unsupervised or semi-supervised techniques could allow to make use of all the available data and provide high-accuracy clusterings and classifications without relying (too much) on domain knowledge.
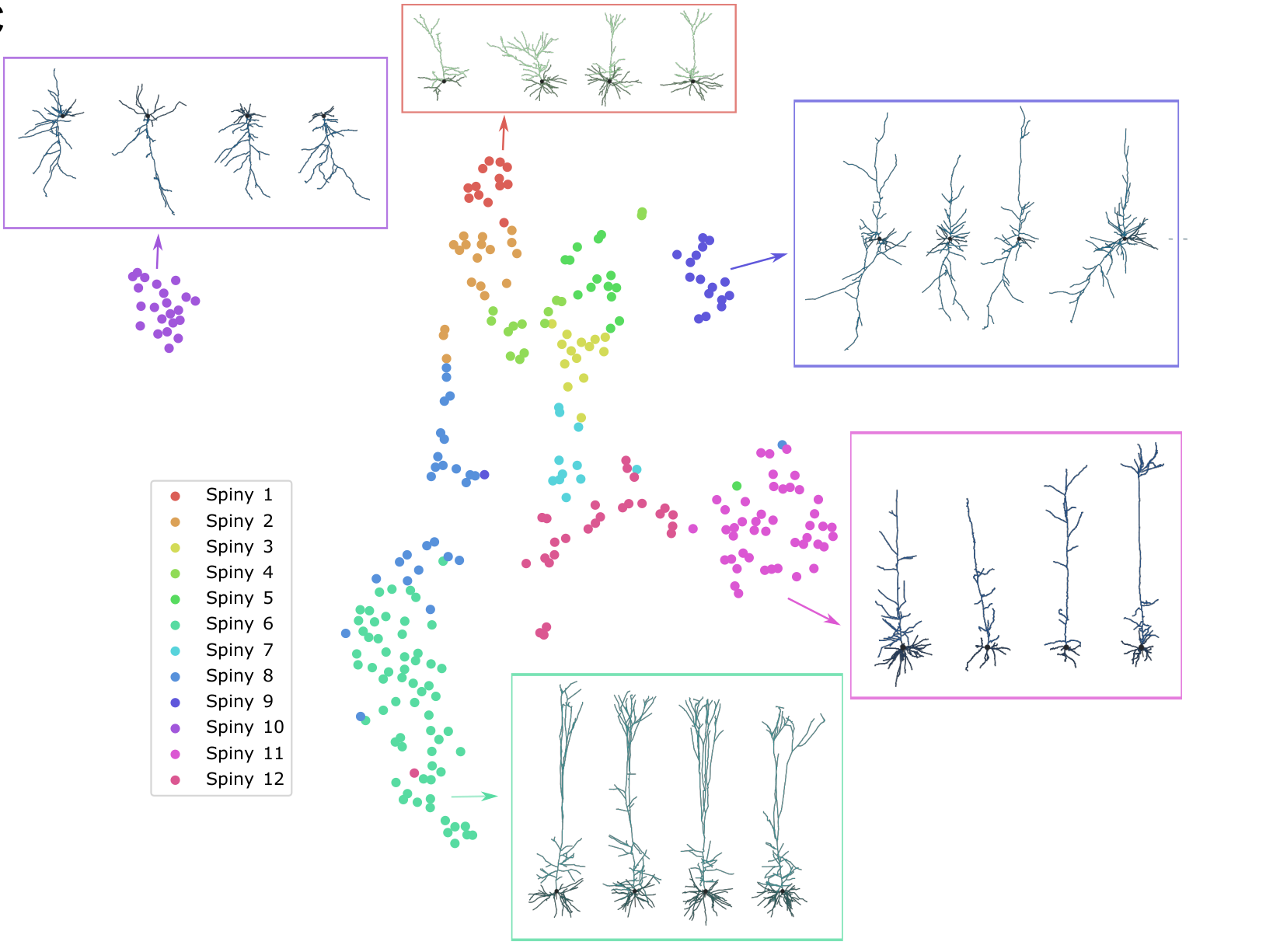
GraphDINO is a contrastive method developed for embedding neurons in order to cluster them using standard clustering methods for vector data. Internally, it uses a combination of a transformer architecture and a classical GNN. In this project, the task is to further develop the message-passing idea within the contrastive architecture. The aim is to develop a model that makes use of deep graph neural networks and/or more expressive GNN architectures and compare that to existing literature.
Requirements
- Python programming
- Interest in working on graph data in 3D
- Interest in neurons and how we can cluster them
- Experience with PyTorch and PyTorch Geometric is a plus
Literature
- Self-supervised Representation Learning of Neuronal Morphologies (“GraphDINO”)
- Graph Representation Learning for Large-Scale Neuronal Morphological Analysis
