Panoptic segmentation of microscopic images of neuronal cell cultures
Using light microscopy to study the behavior of cell cultures is widely used to understand the biological effect of new drug treatments. Thanks to automation, far more microscopic images can now be acquired than any human can possibly analyze manually. This means that computer vision-based analysis is crucial for pushing the field of cell biology forward, and due to the complexity of microscopic images deep learning-based approaches are essential. Sartorius has recently published LIVECell, a large-scale resource to enable deep learning-based instance segmentation of individual cells1. During the preparation of LIVECell, neuronal cell types proved particularly challenging due to their convex morphologies. To address this challenge, Sartorius has put together a specialized dataset of manually annotated images of neuronal cell cultures (see example in the Figure). This neuronal dataset comes with additional non-annotated images, allowing the use of semi-supervised approaches to image segmentation.
The purpose of this project is to use the newly assembled dataset to train and compare deep learning-based models that segment microscopic images of neuronal cell cultures.
- Train a supervised baseline model for panoptic segmentation, meaning simultaneous segmentation of neuronal cell bodies and neurites, on annotated images and evaluate performance using the panoptic quality (PQ)-metric.
- Train at least one model using semi/self-supervised learning leveraging both annotated and non-annotated images and compare it to the supervised baseline model.
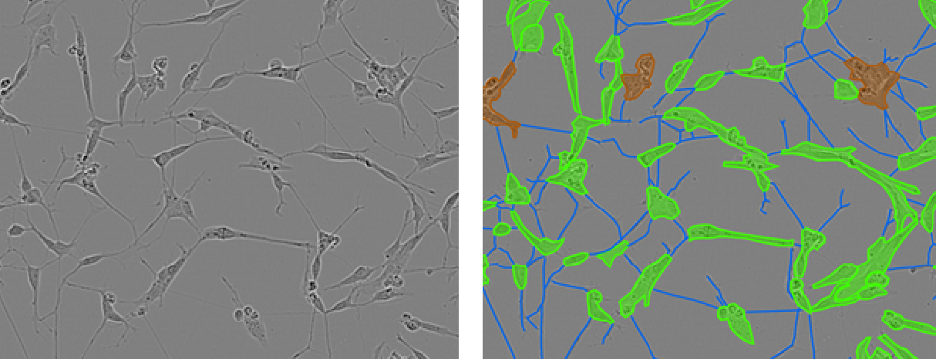
Figure 1: Example phase contrast images of neuronal cell cultures (left) with corresponding annotations overlain (right), where cell bodies are green, neurites are blue and clusters where individual cells can not be separated by eye are brown.
Literature
-
Edlund, C., Jackson, T.R., Khalid, N. et al. LIVECell—A large-scale dataset for label-free live cell segmentation. Nat Methods (2021). https://doi.org/10.1038/s41592-021-01249-6
-
Kirillov, A., He, K., Girshick, R., Rother, C., & Dollár, P. (2019). Panoptic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 9404-9413).
Requirements
- Good mathematical understanding (in particular statistics and linear algebra)
- Python programming
- Experience with deep learning (PyTorch or Tensorflow recommended)
Contact
Rickard Sjögren, Senior Research Scientist, Sartorius Corporate Research. Please CC Fabian Sinz and Alexander Ecker.
