Protein Structure Analysis
The analysis of protein structure has traditionally been a domain restricted to complex and hard-to-use technologies, such as X-ray crystallography or cryo-electron tomography. Fluorescence imaging has never been considered able to analyze such structures, due to its low resolution. This was changed by a recent development termed one-nanometer expansion microscopy (ONE), developed at the University Medical Center Göttingen, in the Institute for Neuro- and Sensory Physiology. This technology relies on expanding biological samples tenfold or even more, followed by a detailed fluorescence fluctuation analysis. The resolution obtained is high enough to show the shape of individual molecules, as seen below.
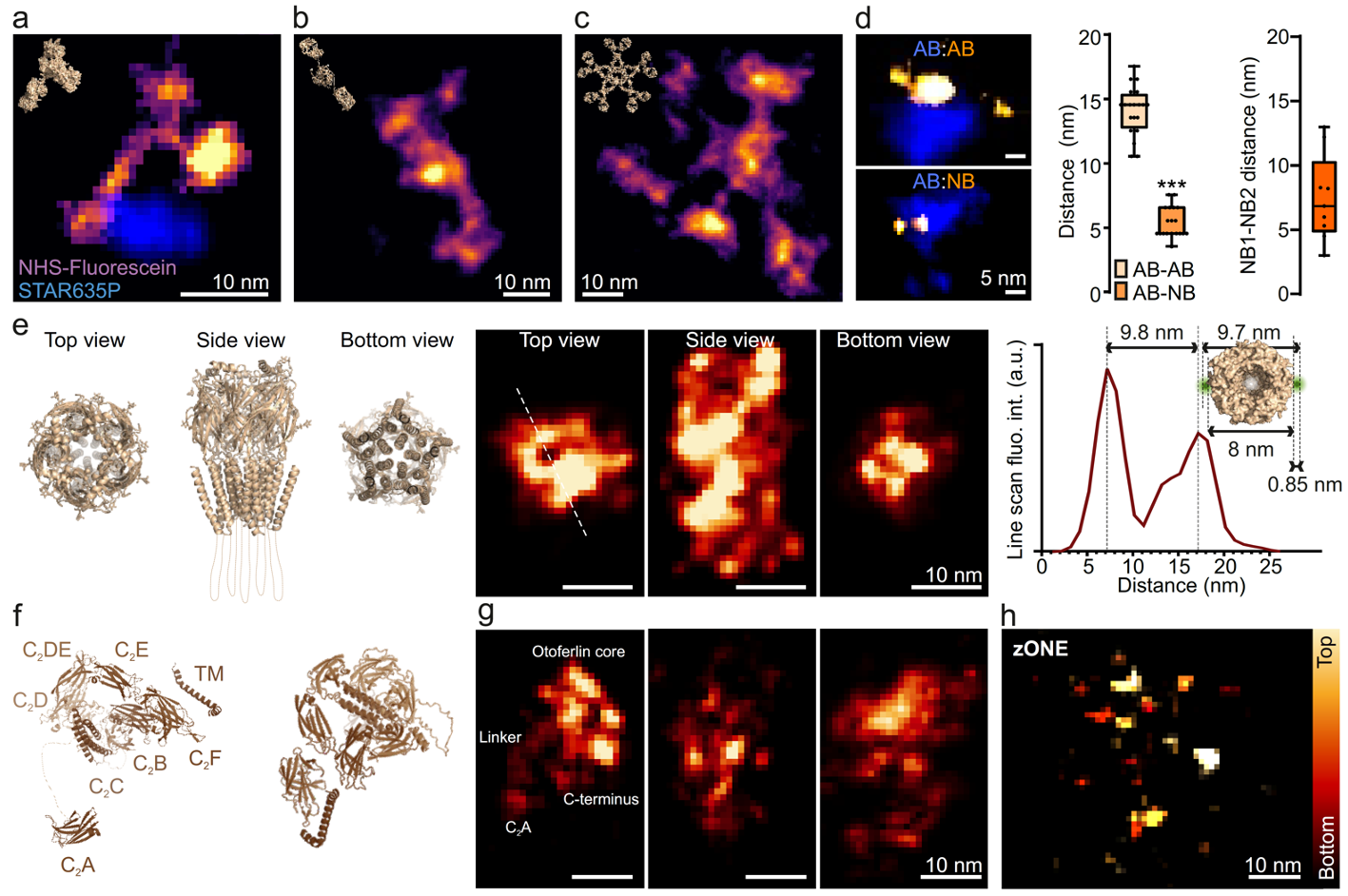
The goal of this project is to go beyond the simple visualization of molecules and to obtain their atomic structure. One challenge of this technique is that the orientation of the sample is random and unknown and therefore needs to be estimated from the data. As a first step in this direction, we will start with proteins of known structure and infer the orientation of each sample using a 3d structural model. Subsequently we will investigate techniques to simultaneously estimate the 3d structure of the protein as well as the view parameters of each individual sample.
This project will enable an analysis of protein structures with cheap and easily available fluorescence microscopes, potentially creating a revolution in the imaging field.
For more details, follow the story on Twitter via #ONE_Microscopy or visit this link.
The ONE microscopy preprint is now available on bioRxiv.
Requirements
- Good mathematical understanding (in particular linear algebra)
- Interest in 3D data
- Python programming
- Experience with gradient-based optimization (e,.g. deep learning frameworks like PyTorch or Tensorflow) would be a plus
